The first capability involves bacterial population profiling for microbiome analyses. Recent studies show that humans harbor vast and diverse populations of microorganisms that are essential to life and health. Research is now focused on correlating health and disease to the composition and changes in an individual’s microbiome. With more than 1,000 publications in 2011, microbiome analysis is one of the fastest growing fields of investigation.
Current techniques for profiling the microbiome in a specimen are labor intensive and can take several weeks. Because Genome Sequence Scanning™ can rapidly detect and quantitate bacterial strains from complex samples, the technology has the potential to become a mainstay of microbiome research.
The second capability involves rapid, high-resolution strain typing. Molecular typing of bacteria is widely used to understand how differences in genomic profile correlate to phenotypic characteristics such as pathogenicity.
Current typing techniques such as pulsed field gel electrophoresis (PFGE) take several days and results are difficult to compare among different operators and laboratories. GSS enables high resolution typing to be performed in hours and with a consistent data format that allows genomic structures to be readily compared.
Chromatin Immunoprecipitation Sequencing
Sunday, January 1, 2017
Can Proteins Interact with DNA and RNA to Influence Nucleic Acid?
Due to the fact that nucleic acids carry genetic information and that proteins regulate various life processes, they are considered to be two of the most important biomolecules in any living organism. In addition, their interactions play a crucial role in most biological processes, which include everything from replication,transcription and recombination to enzymatic eventsusing nucleic acids as substrates. Taking all of these things into consideration, it is not surprising why protein-nucleic acids interactions have been the subject of intensive research for the past few years.
Submit a question and The Protein Man and his team at G-Biosciences will provide you an answer or a resource to aid you. Alternatively other scientists or science enthusiasts can also provide answers, suggestions and comments.
Submit a question and The Protein Man and his team at G-Biosciences will provide you an answer or a resource to aid you. Alternatively other scientists or science enthusiasts can also provide answers, suggestions and comments.
Labels:
Health Life,
Protein,
USA
Wednesday, November 30, 2016
Student need to know How Genomic DNA extraction works?
While there may be a number of ways by which you can successfully isolate your genomic DNA from your sample, your final choice of which genomic DNA extraction protocol to use will ultimately depend on several factors which includes the following:
- the molecular weight of the DNA of interest;
- quantity and purity required to facilitate downstream applications;
- ease or complexity of your chosen method;
- time requirements; and
- budgetary constraints.
Genomic DNA can be separated from all other cellular components by simply following these three basic steps:
Disruption and cell lysis. In extracting your genomic DNA from the sample, you need to break down the cell walls that protect the DNA by using enzymes such as lysozyme and proteinase K or by using physical methods such as manual grinding (mortar and pestle method), freeze-thaw technique, sonication, liquid homogenization and/or mechanical disruption with the use of a Waring blender or a polytron. You can also use bead beating (using 0.1 mm glass beads or 0.15 mm garnet beads) to release your genomic DNA from your cell lysate.
Labels:
Australia,
Health Life,
USA
We need to know How Genomic DNA extraction works?
As mentioned in our previous post, there are a lot of ways by which you can separate your genomic DNA from your sample. Here are some of the most commonly used methods that you can use in extracting genomic DNA from your cell lysate.
NaOH extraction. Known as the "quick-and-dirty" method of preparing DNA, this technique is quite easy to implement and is usually sufficient for most applications. All you need to do is to incubate your cell lysate at high temperatures or subject it to proteinase K digestion and you can have your genomic DNA ready for downstream applications. However, since DNA extracted using this method may contain high levels of contamination, it should not be stored for future use.
Phenol-chloroform extraction. This technique uses organic solvents to extract contaminants from your lysate while the DNA is recovered from the aqueous phase through alcohol precipitation. This may be the most conventional technique of extracting highly purified genomic DNA from a sample but it can also be quite time consuming and may not give reproducible yields.
Silica-based methods. Genomic DNA can easily be extracted from mammalian cells and tissues as well as from mouse tail, E. coli cells and yeast by using silica-based methods. By choosing to use this genomic DNA extraction protocol, you can get your ready-to-use DNA in less than 15 minutes using a spin column based centrifugation procedure. The extracted DNA using this method has an average size of 20 to 30 kb and is ideal for use in PCR, southern blotting analysis and restriction enzyme digestion.
NaOH extraction. Known as the "quick-and-dirty" method of preparing DNA, this technique is quite easy to implement and is usually sufficient for most applications. All you need to do is to incubate your cell lysate at high temperatures or subject it to proteinase K digestion and you can have your genomic DNA ready for downstream applications. However, since DNA extracted using this method may contain high levels of contamination, it should not be stored for future use.
Phenol-chloroform extraction. This technique uses organic solvents to extract contaminants from your lysate while the DNA is recovered from the aqueous phase through alcohol precipitation. This may be the most conventional technique of extracting highly purified genomic DNA from a sample but it can also be quite time consuming and may not give reproducible yields.
Silica-based methods. Genomic DNA can easily be extracted from mammalian cells and tissues as well as from mouse tail, E. coli cells and yeast by using silica-based methods. By choosing to use this genomic DNA extraction protocol, you can get your ready-to-use DNA in less than 15 minutes using a spin column based centrifugation procedure. The extracted DNA using this method has an average size of 20 to 30 kb and is ideal for use in PCR, southern blotting analysis and restriction enzyme digestion.
Labels:
Genomic,
Health Life
Friday, October 28, 2016
Why a microfluidic approach for continuous measurements of biofilm viscosity
The initial measured viscosities for the first 24 hours after inoculation were among the lowest reported to date. Following a low viscosity growth stage, sudden thickening was observed. During this stage, viscosity increased by over an order of magnitude in less than ten hours. The technique was also demonstrated as a promising platform for parallel experiments by subjecting multiple biofilm-laden microchannels to nutrient solutions containing NaCl in the range of 0 mM to 34 mM. Even in this narrow range of ionic strengths, preliminary data suggest a strong relationship between ionic strength and biofilm properties, such as average viscosity and time of onset of rapid thickening. The technique opens the way for a combinatorial approach to study the response of biofilm viscosity under well-controlled physical, chemical and biological growth conditions.
Labels:
Health Life,
Personal Insurance
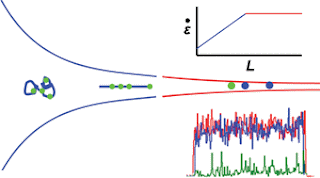
Understanding Constant stretching of DNA in a microfluidic device
The detection is done in a continuous flow microfluidic device with confocal microscopy. In order to carry out the spatial recognition of the fluorescent tags along the length of the DNA fragment, it needs to be stretched out into a linear form using a funnel. High molecule throughput is important as the detection confidence of this technology relies on observing as many tags as possible in the specified experimental period.
The team looks at the relationship between the funnel taper shape and related parameters, such as fluid velocity and fragment length, to improve the current designs and increase throughput. Their new geometries are able to keep the tension applied to the DNA constant during the detection process. Because DNA fragments come in various lengths, a very important goal is to maximise the range of lengths that can be stretched effectively with the funnel. The influence of channel etch depth on fluid flow, and therefore throughput, is also considered.
Labels:
Health Life,
Personal Insurance
Tuesday, October 11, 2016
Understanding What assay development accessories should be used for protein binding?
Protein/Peptide Binding Plates
Plates designed for protein/peptide binding can either be coated with nickel (for binding 6X histidine tagged proteins and peptides), glutathione (for GST tagged proteins and peptides), amine (for binding primary amines of peptides and proteins) and/or sulfhydryl (for free sulfhydryls of peptides and proteins). Most of these plates are supplied as clear, white and black and can ideally be used for colorimetric, chemiluminescent and fluorescent detection methods, respectively.
Nickel coated plates directly isolate polyhistidine-tagged proteins and peptides while glutathione coated plates isolate glutathione S-transferase (GST) tagged proteins from bacterial lysates for ELISA-based protocols. Detergents used to lyse cells do not inhibit protein binding to activated plates as they usually do with plain polystyrene plates. In addition, these plates are ready to use and are pre-blocked to reduce non-specific binding.
Amine coated plates are maleic anhydride activated plates that rapidly bind primary amines of peptides and proteins to form amide bonds that are stable under neutral and basic conditions (pH≥7). Under acidic conditions, however, the bonds will be hydrolyzed releasing the peptide/ligand. Taking this into consideration, binding of peptide/ligand to plates should be performed between pH 8 to 9 and the binding assays or ELISA should be performed at pH conditions ≥7.
Sulfhydryl-binding plates are maleimide activated plates that react with free sulfhydryls to form stable thioether bonds between pH 6.5 and 7.5. These are ideally used in immobilizing and binding sulfhydryl-containing molecules, especially those that are difficult to coat onto polystyrene plates (e.g. protein molecules with free sulfhydryl group and peptides that contain a terminal cysteine). Both amine-binding and sulfhydryl-binding plates are pre-blocked to reduce non-specific binding.
Subscribe to:
Comments (Atom)