The first capability involves bacterial population profiling for microbiome analyses. Recent studies show that humans harbor vast and diverse populations of microorganisms that are essential to life and health. Research is now focused on correlating health and disease to the composition and changes in an individual’s microbiome. With more than 1,000 publications in 2011, microbiome analysis is one of the fastest growing fields of investigation.
Current techniques for profiling the microbiome in a specimen are labor intensive and can take several weeks. Because Genome Sequence Scanning™ can rapidly detect and quantitate bacterial strains from complex samples, the technology has the potential to become a mainstay of microbiome research.
The second capability involves rapid, high-resolution strain typing. Molecular typing of bacteria is widely used to understand how differences in genomic profile correlate to phenotypic characteristics such as pathogenicity.
Current typing techniques such as pulsed field gel electrophoresis (PFGE) take several days and results are difficult to compare among different operators and laboratories. GSS enables high resolution typing to be performed in hours and with a consistent data format that allows genomic structures to be readily compared.
Showing posts with label Health Life. Show all posts
Showing posts with label Health Life. Show all posts
Sunday, January 1, 2017
Can Proteins Interact with DNA and RNA to Influence Nucleic Acid?
Due to the fact that nucleic acids carry genetic information and that proteins regulate various life processes, they are considered to be two of the most important biomolecules in any living organism. In addition, their interactions play a crucial role in most biological processes, which include everything from replication,transcription and recombination to enzymatic eventsusing nucleic acids as substrates. Taking all of these things into consideration, it is not surprising why protein-nucleic acids interactions have been the subject of intensive research for the past few years.
Submit a question and The Protein Man and his team at G-Biosciences will provide you an answer or a resource to aid you. Alternatively other scientists or science enthusiasts can also provide answers, suggestions and comments.
Submit a question and The Protein Man and his team at G-Biosciences will provide you an answer or a resource to aid you. Alternatively other scientists or science enthusiasts can also provide answers, suggestions and comments.
Labels:
Health Life,
Protein,
USA
Wednesday, November 30, 2016
Student need to know How Genomic DNA extraction works?
While there may be a number of ways by which you can successfully isolate your genomic DNA from your sample, your final choice of which genomic DNA extraction protocol to use will ultimately depend on several factors which includes the following:
- the molecular weight of the DNA of interest;
- quantity and purity required to facilitate downstream applications;
- ease or complexity of your chosen method;
- time requirements; and
- budgetary constraints.
Genomic DNA can be separated from all other cellular components by simply following these three basic steps:
Disruption and cell lysis. In extracting your genomic DNA from the sample, you need to break down the cell walls that protect the DNA by using enzymes such as lysozyme and proteinase K or by using physical methods such as manual grinding (mortar and pestle method), freeze-thaw technique, sonication, liquid homogenization and/or mechanical disruption with the use of a Waring blender or a polytron. You can also use bead beating (using 0.1 mm glass beads or 0.15 mm garnet beads) to release your genomic DNA from your cell lysate.
Labels:
Australia,
Health Life,
USA
We need to know How Genomic DNA extraction works?
As mentioned in our previous post, there are a lot of ways by which you can separate your genomic DNA from your sample. Here are some of the most commonly used methods that you can use in extracting genomic DNA from your cell lysate.
NaOH extraction. Known as the "quick-and-dirty" method of preparing DNA, this technique is quite easy to implement and is usually sufficient for most applications. All you need to do is to incubate your cell lysate at high temperatures or subject it to proteinase K digestion and you can have your genomic DNA ready for downstream applications. However, since DNA extracted using this method may contain high levels of contamination, it should not be stored for future use.
Phenol-chloroform extraction. This technique uses organic solvents to extract contaminants from your lysate while the DNA is recovered from the aqueous phase through alcohol precipitation. This may be the most conventional technique of extracting highly purified genomic DNA from a sample but it can also be quite time consuming and may not give reproducible yields.
Silica-based methods. Genomic DNA can easily be extracted from mammalian cells and tissues as well as from mouse tail, E. coli cells and yeast by using silica-based methods. By choosing to use this genomic DNA extraction protocol, you can get your ready-to-use DNA in less than 15 minutes using a spin column based centrifugation procedure. The extracted DNA using this method has an average size of 20 to 30 kb and is ideal for use in PCR, southern blotting analysis and restriction enzyme digestion.
NaOH extraction. Known as the "quick-and-dirty" method of preparing DNA, this technique is quite easy to implement and is usually sufficient for most applications. All you need to do is to incubate your cell lysate at high temperatures or subject it to proteinase K digestion and you can have your genomic DNA ready for downstream applications. However, since DNA extracted using this method may contain high levels of contamination, it should not be stored for future use.
Phenol-chloroform extraction. This technique uses organic solvents to extract contaminants from your lysate while the DNA is recovered from the aqueous phase through alcohol precipitation. This may be the most conventional technique of extracting highly purified genomic DNA from a sample but it can also be quite time consuming and may not give reproducible yields.
Silica-based methods. Genomic DNA can easily be extracted from mammalian cells and tissues as well as from mouse tail, E. coli cells and yeast by using silica-based methods. By choosing to use this genomic DNA extraction protocol, you can get your ready-to-use DNA in less than 15 minutes using a spin column based centrifugation procedure. The extracted DNA using this method has an average size of 20 to 30 kb and is ideal for use in PCR, southern blotting analysis and restriction enzyme digestion.
Labels:
Genomic,
Health Life
Friday, October 28, 2016
Why a microfluidic approach for continuous measurements of biofilm viscosity
The initial measured viscosities for the first 24 hours after inoculation were among the lowest reported to date. Following a low viscosity growth stage, sudden thickening was observed. During this stage, viscosity increased by over an order of magnitude in less than ten hours. The technique was also demonstrated as a promising platform for parallel experiments by subjecting multiple biofilm-laden microchannels to nutrient solutions containing NaCl in the range of 0 mM to 34 mM. Even in this narrow range of ionic strengths, preliminary data suggest a strong relationship between ionic strength and biofilm properties, such as average viscosity and time of onset of rapid thickening. The technique opens the way for a combinatorial approach to study the response of biofilm viscosity under well-controlled physical, chemical and biological growth conditions.
Labels:
Health Life,
Personal Insurance
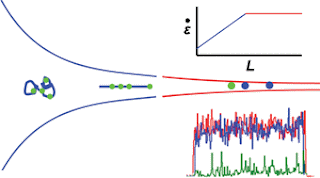
Understanding Constant stretching of DNA in a microfluidic device
The detection is done in a continuous flow microfluidic device with confocal microscopy. In order to carry out the spatial recognition of the fluorescent tags along the length of the DNA fragment, it needs to be stretched out into a linear form using a funnel. High molecule throughput is important as the detection confidence of this technology relies on observing as many tags as possible in the specified experimental period.
The team looks at the relationship between the funnel taper shape and related parameters, such as fluid velocity and fragment length, to improve the current designs and increase throughput. Their new geometries are able to keep the tension applied to the DNA constant during the detection process. Because DNA fragments come in various lengths, a very important goal is to maximise the range of lengths that can be stretched effectively with the funnel. The influence of channel etch depth on fluid flow, and therefore throughput, is also considered.
Labels:
Health Life,
Personal Insurance
Saturday, September 24, 2016
what is Recombinant antibodies?
Both monoclonal and recombinant antibodies can be used in biomedical science and toxicological research, and are effective therapeutic treatments for cancer, autoimmune disorders and a host of other diseases. However, while monoclonal antibodies have become one of the most common tools in biomedical science and medicine due to their ability to bind and neutralize or destroy cell-specific antigens, the ascites method of production causes significant pain and discomfort to the animals used in the process.
As such, the governments of Australia, Germany, the Netherlands and the United Kingdon banned it in favor of in vitro methods. The US also endorses the use of in vitro methods as the default procedure for the production of mAbs.
However, it is important to note that in vitro methods involving hybridomas also have their own limitations, which include the following:
Require immunization and subsequent euthanasia of the animals used in the process.
Slow and labor-intensive.
Often cause immune reactions so the antibodies need to be altered and "humanized" before they can be administered to patients.
Labels:
Health Life,
Personal Insurance
Understanding between Monoclonal vs Polyclonal Antibodies
Monoclonal antibodies (mAbs) represent a population of antibodies that recognize a single epitope within an antigen. Since mAbs are produced from a single B cell in the spleen or lymph nodes of an immunized mouse, the resulting antibodies are all identical. In addition, they recognize the same epitope of a specific antigen.
However, while B cells can be used to harvest antibodies, these cells have a limited lifespan and will eventually stop producing the antibody in time. To overcome this limitation, a specific antibody-producing B cell is fused with a myeloma cell to create an immortalized B cell-myeloma hybridoma which can provide a constant supply of highly specific monoclonal antibody.
Monoclonal antibodies can be raised against many targets. Specific antibody characteristics (sensitivity requirements and cross reactivity levels) can be identified and monoclonal antibodies screened to identify any cell lines exhibiting the required characteristics.
Monoclonals can also be generated to cross react with a group of molecules. This can be quite useful in cases where there are multiple possible combinations of drugs to be tested in a patient.
Monoclonals are typically rat or mouse monoclonals, but they can also be generated from various species such as rabbit and goat.
Advantages of mAbs
Immortal supply. Once a monoclonal antibody hybridoma is created, it can produce unlimited quantities of highly specific antibodies.
High specificity. mAbs are ideal for use as primary antibody in an assay, or for detecting antigens in tissues.
High reproducibility. They produce low background staining and lot-to-lot variation, and have low cross-reactivity. As such, they provide reproducible results and ensure efficiency in affinity purification.
Labels:
Health Life
Saturday, August 6, 2016
You need to know How do you prepare your phosphate buffer?
Preparing Your Phosphate Buffer
How do you prepare your phosphate buffer? Here's how:
What You'll Need
Monosodium phosphate, monohydrate and its conjugate base disodium phosphate, heptahydrate
Water
Phosphoric acid (to make the buffer more acidic) or sodium hydroxide (to make the solution more basic)
pH meter, glassware and hot plate with stirring bar
Procedure:
Decide on the properties of your buffer. Before doing anything, you need to decide on the molarity and pH of your buffer. Remember, most buffers are used at a concentration between 0.1 M and 10 M and the pH should ideally be within 1 pH unit of the acid or its conjugate base.
Prepare the solution. Using the calculated ratioderived from the equation, mix the required amounts of acid and base and mix them in approximately one liter of water to prepare a rough buffer solution.
Check the pH. Use your pH meter and adjust the pH accordingly by using phosphoric acid or sodium hydroxide. Bring the total volume to one liter once you have reached the desired pH.
Dilute as necessary. Use this stock solution to prepare buffers of different molarities as needed.
Labels:
Health Life,
Personal Insurance
Understanding SCANNING GENOMIC INFORMATION AND INNOVATION
Shiga-toxigenic Escherichia coli (STEC) and Salmonella enterica are an important U.S. public health concern, causing an estimated 1.2 million cases of foodborne illness each year. E. coli O157:H7, the most common of the virulent STECs, has been implicated in multiple foodborne illness outbreaks, and six additional STEC serogroups are now considered adulterants in certain beef products. Unlike STEC O157:H7, however, the “Big 6” virulent strains are not easily distinguished using available molecular testing methods.
Of the more than 2500 identified strains of Salmonella enterica, 1700 of them are classified as human pathogens belonging to subspecies S. enterica I. Just 20 of these serotypes are responsible for greater than 70% of the illnesses caused by S. enterica subspecies I.
Clearly there is a need to be able to rapidly and cost-effectively identify these more virulent strains when present in food products.
PathoGenetix’s GSS technology identifies microbial DNA from complex mixtures or from isolates, and automates the process from sample preparation through data analysis to provide actionable information in five hours. Because GSS scans microbial DNA directly from a mixed culture and does not require a pure culture, it can reduce the time, complexity, skill and cost required for molecular identification and strain typing. br />
Labels:
Health Life
Tuesday, July 12, 2016
Understanding PathoGenetix Delivers Bacterial Identification System to FDA
The RESOLUTION System, based on PathoGenetix’s proprietary Genome Sequence Scanning™ (GSS™) technology, enables pathogen serotype identification and strain typing in just five hours, directly from complex mixtures such as enriched food and clinical samples. The bacterial strain information provided by the RESOLUTION System is comparable to pulsed field gel electrophoresis (PFGE), the current gold standard for pathogen typing in foodborne illness outbreak investigation and response.
Identifying the pathogen strain that is causing a foodborne illness outbreak is a critical step in defining the extent of the outbreak, determining the food involved, finding the original source of the contamination and defining the scope of a product recall. The ability of the RESOLUTION System to derive useful pathogen strain and serotype information directly from a complex mixture, and to shorten the time for pathogen typing to just five hours, could allow for quicker decisions affecting public health.
Current microbial identification techniques such as PFGE and whole genome sequencing (WGS) require a cultured isolate as input, and advanced, time-consuming laboratory processes for preparation and processing of food samples. Analysis of the patterns created by PFGE, or the extensive data generated by WGS, can be complex and add significantly to the time required to identify the pathogen strain and serotype. Because the PathoGenetix system is culture independent, and fully automated from sample preparation to final report, it has the potential to greatly reduce the time, complexity and skill-level required to identify foodborne pathogens in hospital and public health labs monitoring foodborne outbreaks.
Labels:
Business Insurance,
Health Life,
Personal Insurance
What is Genome Sequence Scanning (GSS) technology
The BioNumerics® software suite (Applied Maths) was used to analyze a data set of 190 pathogenic E. coli strains from the Centers for Disease Control and Prevention (CDC). Clustering of related strains was performed using patterns generated by PFGE and whole genome sequence data included in the CDC data set, and GSS fingerprints, PathoGenetix’s proprietary technology used in the RESOLUTION™ Microbial Genotyping System. For the set of E. coli isolates tested, the analysis shows a remarkably high congruence between the GSS groupings and WGS groupings, while maintaining a good concordance with the PFGE groupings. With respect to WGS, the GSS groupings also turn out to be more discriminatory than the PFGE groupings.
The RESOLUTION System can work from a mixed sample and does not require the preparation of a cultured isolate, as is the case with whole genome sequencing and PFGE, and provides strain type and serotype results in less than five hours.
The collaborative research is detailed in a poster presented yesterday at the InFORM 2013 meeting being held this week in San Antonio, Texas. InFORM meetings are designed to coordinate and enhance the work of microbiologists, epidemiologists and environmental health specialists focused on foodborne disease surveillance, outbreak detection and response. The meeting is sponsored by the CDC, the Association of Public Health Laboratories (APHL), the U.S. Department of Agriculture Food Safety and Inspection Service (FSIS), and the Food and Drug Administration (FDA), and integrates the separate PulseNet and OutbreakNet annual meetings held in previous years.
Applied Maths and PathoGenetix signed a collaborative agreement in June to integrate the RESOLUTION Microbial Genotyping System with the BioNumerics software suite. The two companies have a first working version of the new functionality ready, which integrates PathoGenetix’s rapid pathogen strain typing with BioNumerics’ advanced data management and networking tools. The plug-in will enable sharing and comparison of outbreak strain data among public health or food safety testing labs, and the comparison of serotype and strain type information generated by the RESOLUTION System with data sets generated by other identification methods such as PFGE or whole genome sequencing.
Labels:
Health Life
Tuesday, June 28, 2016
We need to know Why Is Lactate Dehydrogenase (LDH) Release A Good Measure For Cytotoxicity?
On the other hand, cells undergoing apoptosis (normal or programmed cell death) go through a series of well-defined events such as the shrinking of the cytoplasm, cleavage of DNA into smaller fragments, etc. before being engulfed by white blood cells.
When the cell membranes are compromised or damaged in any way, lactate dehydrogenase (LDH), a soluble yet stable enzyme found inside every living cell, is released into the surrounding extracellular space. Since this only happens when cell membrane integrity is compromised, the presence of this enzyme in the culture medium can be used as a cell death marker. The relative amounts of live and dead cells within the medium can then be quantitated by measuring the amount of released LDH using a colorimetric or fluorometric LDH cytotoxicity assay.
Other enzymes such as adenylate kinase and glucose-6-phosphate may also be used to measure cytotoxicity but these enzymes are not stable and lose their activity during cell death assays.
Labels:
Business Insurance,
Health Life
Understanding How Are They Classified?
In general, there are three distinct types of cytotoxicity assays. There are assays that determine cell viability by:
- Exhibiting a change in the membrane permeability or metabolism (viability assays);
- Measuring their absolute long term survival rate and their capacity to regenerate (long term survival assays);
- Exhibiting survival in an altered or genetically mutated state (irritancy assays).
By using a silicon microphysiometer, reduction in the extracellular acidification rate as a result of any changes in the intracellular metabolism can be detected.
Since each of these approaches has its own advantages and disadvantages, it is highly recommended that you use a variety of cytotoxicity assays to accurately determine cell viability.
Labels:
Canada,
Health Life
Monday, June 20, 2016
Understanding How to extract biologically active proteins from the cells
- Expensive
- Cumbersome due to use of heavy equipment
- Reduced yield as the sample is processed through several steps
- Batch to batch variation in protein yield due variability and handling
Mechanical methods can sometimes denature proteins as these methods are harsh and some of them generate heat and can denature protein if appropriate cooling of sample is not done.
Chemical methods for cell disruption using lysis buffers with ionic detergents can be used to release proteins; however they can denature the protein. Non-ionic detergents offer advantage over lysis buffer and ionic detergents as they are mild and the proteins are not denatured. Therefore, nowadays popular methods for extraction of biologically active proteins from cells or tissues use non-ionic detergents along with other additives, such as enzymes for cell wall disruption and addition of protease inhibitors to inhibit proteases. This method may involve mild mechanical methods, including brief homogenizing or vortexing depending upon the cell or tissue type.
Labels:
Business Insurance,
Health Life,
Personal Insurance
Do you know What Detergents Used in Proteomics?
When the detergent is introduced into the lysate, its molecules insert themselves in the lipid membrane and begin separating the lipid bi-layer. As the detergent's concentration increases, the lipid bi-layer becomes saturated with detergent molecules and then starts breaking apart producing protein-detergent complexes and detergent-lipid micelles in the process.
It is interesting to note that while detergents start to form highly organized spherical structures (micelles)in aqueous solutions, as it reaches its Critical Micelle Concentration (CMC), they tend to form reverse micelles in non-aqueous solutions and/or in the presence of hydrocarbon solvents instead.
In general, detergents can be classified as follows:
Ionic.Ionic detergents are ideally used for completely disrupting the cellular structure as well as for denaturing a wide variety of proteins for separation during gel electrophoresis. Some of the most commonly used ionic detergents include the anionic detergents sodium dodecyl sulfate (SDS) and deoxycholate, and the cationic detergent hexadecyltrimethylammonium bromide(CTAB).
Non-ionic.Non-ionic detergents have an uncharged hydrophilic head group and are usually based on polyoxyethylene and/or glycoside. Due to their non-denaturing properties, these detergents are commonly used in isolating biologically active membrane proteins and are most effective in breaking lipid-lipid and lipid-protein interactions. The Triton, Tween and Brij series are great examples of polyoxyethylene-based non-ionic detergents while octyl beta-glucoside and the MEGA series detergents are perfect examples of glycoside-based non-ionic detergents.
Labels:
Health Life
Friday, June 10, 2016
So why PathoGenetix Announces $10 Million Series C Financing
The GSS technology confirms and identifies microbial DNA in either complex mixtures or isolates, and provides actionable information in five hours. The funding will be used to further develop the first commercial system using the GSS technology, the RESOLUTION™ Microbial Genotyping System. The fully automated RESOLUTION System includes the instrument, bioinformatics software and database, and pathogen-specific assays, and will be commercially available in 2014 for use in food safety testing and foodborne illness outbreak investigations.
“This commitment from our investors reflects the outstanding progress we have made toward commercialization of the GSS technology," said Ann Merrifield, President and CEO of PathoGenetix. "As we continue our efforts to deliver the RESOLUTION System for rapid pathogen strain typing, we also are gaining excellent market traction in the key public health and food industry sectors.”
The company announced collaborations with the Food and Drug Administration (FDA) in April and the U.S. Department of Agriculture’s Agricultural Research Service (USDA-ARS) in September, and is currently conducting a series of customer evaluations with leading food safety testing labs. In June, the company signed an agreement with Applied Maths, NV, to link the RESOLUTION System with the BioNumerics software suite currently in use in thousands of public and private research sites and laboratories around the world.
The company announced collaborations with the Food and Drug Administration (FDA) in April and the U.S. Department of Agriculture’s Agricultural Research Service (USDA-ARS) in September, and is currently conducting a series of customer evaluations with leading food safety testing labs. In June, the company signed an agreement with Applied Maths, NV, to link the RESOLUTION System with the BioNumerics software suite currently in use in thousands of public and private research sites and laboratories around the world.
Labels:
Health Life,
Personal Insurance
Wednesday, June 1, 2016
Undertanding Present DNA Stretching Technology at APS Physics Meeting
PathoGenetix’s Genome Sequence Scanning (GSS) is a bacterial identification technology that detects sequence-specific fluorescent tags on long DNA molecules that have been extracted and purified directly from biological samples. Key to the efficacy and sensitivity of the GSS technology is the microfluidic funnel used for high-throughput stretching and scanning of long strands of single DNA molecules. In the proprietary GSS detection funnels, purified and tagged DNA molecules flow in a linear conformation at high speed past a series of lasers and optical sensors, which record the length and pattern of the labels on each DNA fragment. The labels create a barcode of the DNA in the sample, which is compared to an onboard database of barcodes to identify the serotype/strain type for the organism.
Labels:
Health Life,
USA
Tuesday, May 24, 2016
Understanding PathoGenetix to Present DNA Stretching Technology
The GSS technology has broad applicability in food safety, industrial microbiology, public health and research, and will be available in the RESOLUTION™ Microbial Genotyping System in 2014 for use in food safety testing and foodborne illness outbreak investigations. Because Genome Sequence Scanning is culture independent, and fully automated from sample preparation to final report, the technology greatly reduces the time, complexity and skill required when compared to other molecular and next generation sequencing (NGS) identification approaches. The strain-type information provided by GSS is comparable to pulsed field gel electrophoresis (PFGE), the current standard for pathogen typing in foodborne outbreak investigation and response. As a result, GSS offers a powerful new tool for epidemiological investigations and outbreak monitoring in hospital and public health labs monitoring foodborne outbreaks, and for food safety testing in the food industry.
Labels:
Health Life
Saturday, May 21, 2016
Understanding between DNA Purification in Ethanol vs. Isopropanol?
Ethanol is usually the solvent of choice when it comes to precipitating DNA out of a solution but you can also use isopropanol and basically get the same results in the end. So, why do some people use ethanol while some prefer to use isopropanol? What is the difference between the two and how do you know which solvent touse for DNA purification? Here are some things you need to know to help you choose the most suitable solvent for your experimentations.
As mentioned earlier, you can use ethanol or isopropanol in precipitating DNA from the solution and get the same end results. However, the solubility of DNA differs in each of these solvent. For the record, DNA is less soluble and falls out of the solution faster even when low concentrations of isopropanol are used but there is a tendency that the salt will co-precipitate with the DNA.On the other hand, a higher concentration of ethanol is needed to precipitate DNA from the solution but then the salts tend to stay soluble, even at lower temperatures.
So, which solvent should you use? Well, this may depend on the volume of the sample you need to precipitate. As a general rule, you should use ethanol if you are only precipitating small volumes of DNA. This way, you can recover larger volumes of DNA without worrying about salt contamination as you would when you choose to use isopropanol. You should also use ethanol if your sample will be stored for long periods of time under low temperatures.
Isopropanol should be used when working with larger samples since you would only need a small amount of the solvent. However, you need to precipitate with minimal incubation time and carry it out at room temperature to minimize salt precipitation.
Labels:
Health Life
Subscribe to:
Posts (Atom)